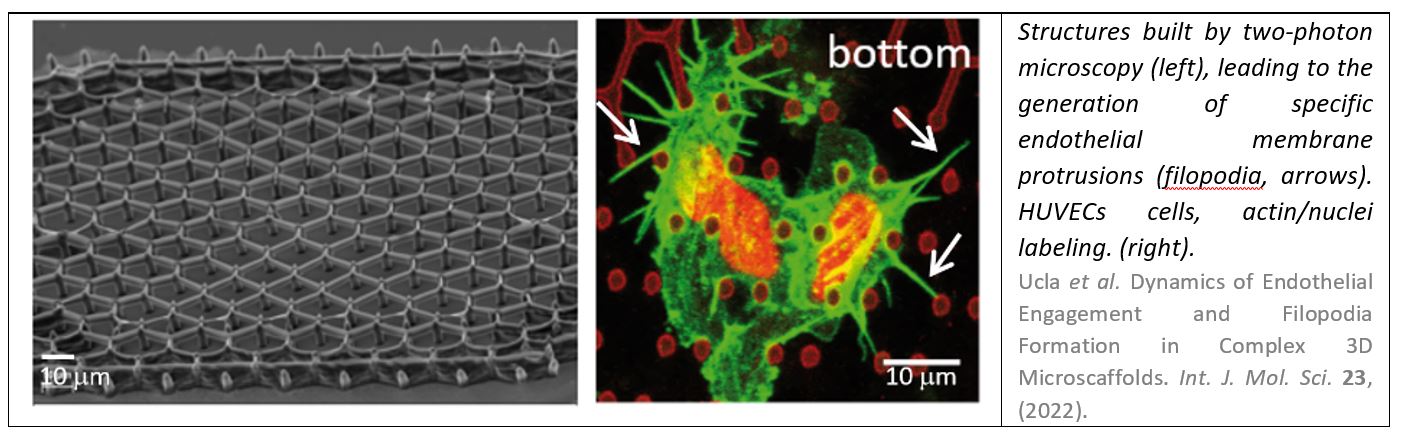
The mechanical, geometrical and chemical properties of the 3D fibrous extracellular matrix play a key role in cancer initiation and progression, by controlling the movement and fate of tumor cells as well as surrounding fibroblasts, immune or endothelial cells. A strong emphasis has been put recently on the development of matrices with controllable fiber properties as tools to understand cell mechanical behavior and migration. However, while numerous biomaterials have been engineered to obtain the desired properties at the multicellular scale, controlling the 3D architecture and the chemistry of fibers at a subcellular scale necessitates specific techniques like two-photon polymerization. Our team has developed over the years a full toolbox to produce at will complex 3D geometries and fiber networks, in hybrid structures made from polymers, hydrogels or protein materials, which also includes an innovative measurement of traction forces exerted by cells. Our system provides now an ideal tool to understand finely the dynamic adaptation of cell individual and collective migration to the microenvironment, and in particular fundamental aspects of cancer cell behaviors.
This internship will aim to characterize the migratory behavior of tumor cell lines from breast and renal cancers, in relation to the fine geometry and local mechanical properties of their 3D fiber microenvironments. Two-photon polymerization will be used to build a variety of generic architectures of fiber grids characteristic of the tumor extracellular matrix, like structures with gradients of stiffness or fiber density. The migration and the dynamic changes of morphology of fluorescently labeled cancer cells will be assessed on these structures, in link with the local physical and chemical cues of the grid. In particular, cancer cells are expected to experience different motilities and morphologies compared to related non-cancer cells. These global changes are related to finer differences at a subcellular scale, for example membrane protrusive states characteristic of cancer cells. An important part of this internship will be to develop from previous tools of the group, including using Artificial Intelligence (Convolution Neural Networks), and from the existing literature image analysis pipelines to characterize these 3D shape changes, and to integrate these results in a general frame.
The internship will have an important focus on the development of 3D image analysis tools dedicated to the project. The aim will be to characterize cell migratory properties in 3D fiber grids, as well as the global morphology and the formation of membrane protrusions. Characteristic outputs will be migration speed, nuclei deformation, global shape changes, dynamics of protrusion formation, and exerted forces. The intern will also be trained to microfabrication by two-photon polymerization, cell culture and advanced optical microscopy – spinning disk, Lattice Light Sheet microscopy. Sets of cancer cell movies will be realized in the different architectures and chemistries. The project will benefit from the rich infrastructure of Institut Curie and Chimie Paris (state-of-the-art imaging platform, cell culture, microfluidics and microfabrication, modeling of biophysical phenomena, wide expertise in chemistry), with a two-photon polymerization set-up specifically dedicated to the project.
- Ucla, P. et al. Dynamics of Endothelial Engagement and Filopodia Formation in Complex 3D Microscaffolds. Int. J. Mol. Sci. 23, (2022).
- Coscoy, S. et al. Microtopographies control the development of basal protrusions in epithelial sheets. Biointerphases 13, 041003 (2018).
- Mukherjee, A. et al. Quantitative biophysical metrics for rapid evaluation of ovarian cancer metastatic potential. Mol. Biol. Cell 33, ar55 (2022).
- Castilla, C., Maska, M., Sorokin, D. V, Meijering, E. & Ortiz-de-Solorzano, C. Three-Dimensional Quantification of Filopodia in Motile Cancer Cells. IEEE Trans. Med. Imaging 1–1 (2018). doi:10.1109/TMI.2018.2873842
- Driscoll, M. K. et al. Robust and automated detection of subcellular morphological motifs in 3D microscopy images. Nat. Methods (2019). doi:10.1038/s41592-019-0539-z
- Poincloux, R. et al. Contractility of the cell rear drives invasion of breast tumor cells in 3D Matrigel. Proc. Natl. Acad. Sci. U. S. A. 108, 1943–1948 (2011).
- Huna, A. et al. Loss of the metastasis suppressor nme1, but not of its highly related isoform nme2, induces a hybrid epithelial–mesenchymal state in cancer cells. Int. J. Mol. Sci. 22, (2021).
- Wang, X. et al. Multi-Omics Profiling to Assess Signaling Changes upon VHL Restoration and Identify Putative VHL Substrates in Clear Cell Renal Cell Carcinoma Cell Lines. Cells 11, (2022).
- Chen, B. C. et al. Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science 346, (2014).